Data use
DEAP offers seven file formats for your custom datasets making it flexible to your preferred data analysis tools and packages.
It is recommended that you use parquet format for tabulated data given its encoded data information (i.e. data type), compression efficiency, and faster load times.
CSV & TSV
Plain text formats (TSV/CSV) are widely compatible and easy to inspect, but less efficient for large datasets. They don’t support selective column loading or preserve metadata, such as data type specification; the metadata is instead available via the sidecar JSON files for plan text files. As a result, tools like Python or R must guess data types during import, often incorrectly. For example, categorical values like “0”/“1” for “Yes”/“No” (commonly used in NBDC datasets) may be interpreted as numeric, and columns with mostly missing values may be treated as empty if the first few rows lack data.
To avoid such issues, it’s recommended to manually define column types using the accompanying data dictionaries included in the sidecar JSON metadata files during the import. The NBDCtools R package offers a helper function, read_dsv_formatted(), to automate this process.
Stata
Due to character limits Stata files use the column name name_stata instead of the standard variable. Stata (and SPSS) is also not available for use in the pre-assembled datasets due to their restrictions on final size; utilize custom datasets instead.
Due to character limits Stata & SPSS files use the column name name_short instead of the standard variable. Stata & SPSS are also not available for use in the pre-assembled datasets due to their restrictions on final size; utilize custom datasets instead.
Parquet
Please note that Parquet files are not officially supported by the BIDS specification. For NBDC datasets, we decided to add Parquet as an alternative file format to the BIDS standard TSV/sidecar json pairs to allow users to take advantage of the features of this modern and efficient open source format that is commonly used in the data science community.
Apache Parquet is a modern, compressed, columnar format optimized for large-scale data. In contrast to TSV files, Parquet supports selective column loading and smaller file sizes. This improves loading speed and memory usage and enhances performance for analytical workflows.
Crucially, parquet can store metadata (including column types, variable/value labels, and categorical coding) directly in the file (as opposed to a sidecar json), enabling accurate import without manual setup. See details for how Parquet export is handled in Lasso and DEAP.
polars module:
import polars as pl
parquet_df = pl.read_parquet("path/to/file.parquet")pandas module:
import pandas as pd
parquet_df = pd.read_parquet("path/to/file.parquet")arrow package:
library(arrow)
parquet_df <- read_parquet("path/to/file.parquet")_df = pd.read_parquet("path/to/file.parquet")Shadow Matrices (HBCD only)
Shadow matrices are currently only available for the HBCD dataset. For ABCD upon download you can select how you would like missigness handled, coded or NULL.
Each TSV and Parquet data file in the BIDS /rawdata/phenotype/ directory has a corresponding shadow matrix file in the same format (TSV or Parquet). These shadow matrix files mirror the structure and column names of the original data files.
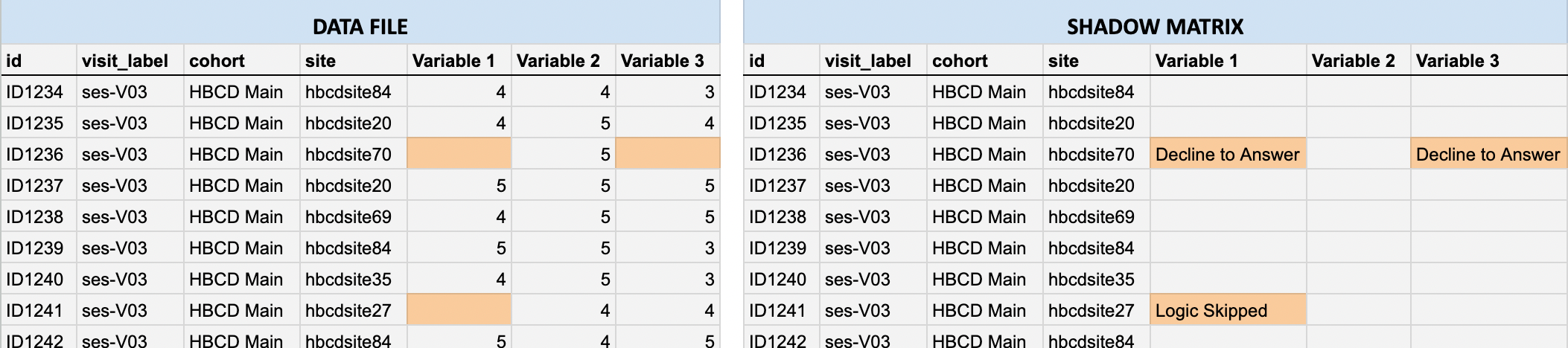
In the data files, missing values are represented as blank cells. Shadow matrices provide essential context by indicating the reason a value is missing (e.g., Don’t know, Decline to answer, Missed visit). Each cell in a shadow matrix corresponds to the same cell in the associated data file: If a data cell contains a value, the corresponding shadow matrix cell is blank.
If a data cell is missing, the corresponding shadow matrix cell includes a code or description indicating the reason, as illustrated below by the highlighted cells in the data file (left) vs. the corresponding shadow matrix (right).
In HBCD, some participant responses like Don’t know or Decline to answer (which are typically considered non-responses) are deliberately converted to missing values in the data file, with the original response converted to a missingness reason stored in the shadow matrix. This prevents analytical errors such as inadvertently treating placeholder codes (like 777 or 999, common in other datasets) as valid numeric values during analysis and ensures consistency in data types across all entries (e.g. text notes in numeric fields are avoided).
Shadow matrices usefullness
While the approach of storing missingness reasons in a shadow matrix file supports cleaner analyses, there are situations where non-responses are themselves meaningful. For example, a researcher might be interested in how often participants do not understand a given question and how this relates to other variables. In such cases, users can re-integrate the non-responses from the shadow matrix back into the data.
Utilizing shadow matrices (R & Python)
Here we describe how researchers can use shadow matrix files in combination with the data files to, for example, explore and understand patterns of missing data or integrate missingness reasons (e.g., Decline to Answer, Logic Skipped, etc.) into your analysis.
For working in R, we recommend using the NBDCtools package - see details here. For Python, the following helper function joins the tabulated data file with its corresponding shadow matrix file so data columns are combined with columns providing the reasons for missingness in the same data frame. This function works with both TSV and CSV file formats, but can be updated for Parquet files using the loading logic shown under the section on Parquet files above.
import pandas as pd
import os
def load_data_with_shadow(data_path, shadow_path):
"""
Loads a data file (CSV or TSV) and its corresponding shadow matrix
(CSV or TSV) and adds '_missing_reason' columns for missing values.
"""
# Detect delimiter from file extension and load data
def get_delimiter(path):
ext = os.path.splitext(path)[1].lower()
return "\t" if ext == ".tsv" else ","
data = pd.read_csv(data_path, delimiter=get_delimiter(data_path))
shadow = pd.read_csv(shadow_path, delimiter=get_delimiter(shadow_path))
# Annotate data with non-empty missingness reason columns (excluding participant_id
# and session_id) in shadow matrix
for col in data.columns[2:]:
if col in shadow.columns:
if not shadow[col].isna().all() and not (shadow[col] == '').all():
data[f"{col}_missing_reason"] = shadow[col]
return data
# Example usage:
df = load_data_with_shadow("data.tsv", "shadow_matrix.tsv", save=True)
# Example: View reasons for missing data for a given column/variable in the data file
df[df["<COLUMN NAME>"].isna()][["<COLUMN NAME>_missing_reason"]]